
AutoStructure is a rule-based expert system, which automatically determines protein structures from NMR data using various analysis rules generalized from basic procedures developed by human experts. AutoStructure generate a reliable protein fold using intelligent analysis methods based on spectrum-specific properties and the iterative identification of self-consistent NOE contact patterns, without using any 3D structure model.The software identifies secondary structures, including alignments between beta-strands, based upon the combine pattern analysis of secondary structure specific NOE contacts, chemical shift, scalar coupling constant, and slow amide proton exchange data.
The software generates conformational constraints (e.g. distance, dihedral angle and hydrogen bond constraints) automatically and submits parallel DYANA structure calculations to an array of Pentium III processors. The resulting structure is then automatically-refined by iterative cycles of assigning self-consistent NOESY cross peaks and regeneration of the protein structure with parallel DYANA calculation.
Coming soon ...
Y.J. Huang et at. Automated determination of protein structures from NMR data by iterative analysis of self-consistent contact patterns (In preparation)
AutoStruture-1.0AutoStruture-1.1 (last updated Oct. 02, 2002)
Protein Structures determined using AutoStructure
FGF-2 (155 a.a.) Basic fibroblast growth factor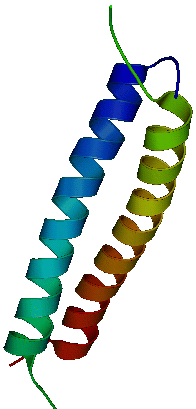
1IHQGlytm1Bzip (38 a.a. in each chain) A Chimeric Peptide Model Of The N-Terminus Of A Rat Short Tropomyosin With The N-Terminus Encoded By Exon 1BGreenfield et al, J Mol. Biol. (2001) 312:833-847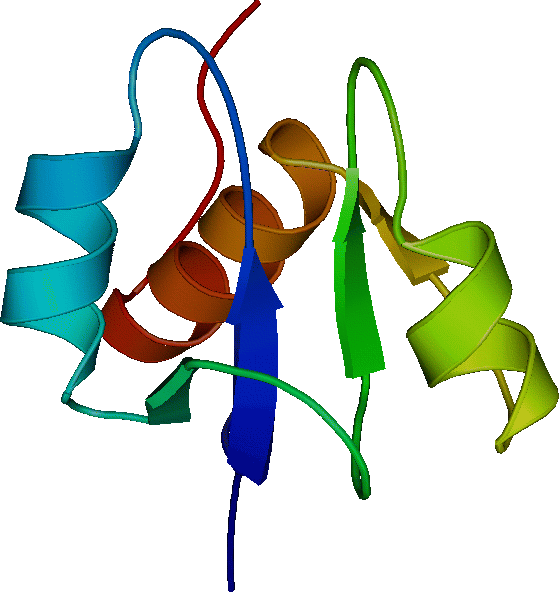
1L7BWR64Tt (92 a.a.) - NESG target 
1KKGWR90Ec (108 a.a.) - NESG target RNA binding protein