- COPY
- [SELEction=<selection>] copies main coordinate
set into comparison set ;
XCOMP
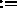 X,YCOMP
X,YCOMP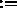 Y,ZCOMP
Y,ZCOMP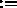 Z; B,Q are
unaffected (default for selection: (ALL) ).
Z; B,Q are
unaffected (default for selection: (ALL) ).
- FIT
- { [SELEction=<selection>]
[MASS=<logical>]
[LSQ=<logical>] } rotates (if LSQ is TRUE) and translates all main coordinates to obtain a best fit between the selected main and comparison atoms. Translation superimposes the geometric centers (or the centers of mass if MASS is TRUE) of the two coordinate sets. Rotation occurs with the Kabsch (1976) least-squares fitting algorithm. Mass-weighting is applied if MASS is set to TRUE. Upon successful completion of this operation, the Eulerian angles describing the rotation matrix R are stored in the symbols $THETA1, $THETA2, $THETA3, and the translation vector T is stored in the symbols $X, $Y, $Z. The fitted coordinate set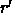 is
related to original set r by
is
related to original set r by 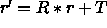 (default for
selection: (ALL); for MASS: FALSE; for LSQ: TRUE).
(default for
selection: (ALL); for MASS: FALSE; for LSQ: TRUE).
- FRACtionalize
- { [SELEction=<selection>]
[A=<real>] [B=<real>] [C=<real>]
[ALPHa=<real>] [BETA=<real>] [GAMMa=<real>] } fractionalizes selected coordinates. The X-PLOR convention keeps the direction of the x-axis (x same direction as a; y is in (a,b) plane) and is the same that is used internally by all X-PLOR routines (default for selected atoms: (ALL); for a, b, c: 1.0; for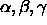 : 90
: 90 ).
).
- INITialize
- { [SELEction=<selection>] } initializes main coordinate set; i.e., X:=9999.0, Y:=9999.0, Z:=9999.0; B,Q are unaffected (default for selection: (ALL) ).
- ORIEnt
- { [SELEction=<selection>]
[MASS=<logical>]
[LSQ=<logical>] } rotates (if LSQ is TRUE) and translates all coordinates such that the principal axis system of the selected atoms corresponds to the x,y,z axis. The translation superimposes the geometric center (or the center of mass if MASS is TRUE) and the coordinate origin. The rotation occurs with the Kabsch (1976) least-squares fitting algorithm. Mass-weighting is applied if MASS is set to TRUE. Upon successful completion of this operation, the Eulerian angles describing the rotation matrix R are stored in the symbols $THETA1, $THETA2, $THETA3, and the translation vector T is stored in the symbols $X, $Y, $Z. The oriented coordinate set is
related to original set r by
is
related to original set r by 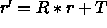 (default for
selection: (ALL); for MASS: FALSE; for LSQ: TRUE).
(default for
selection: (ALL); for MASS: FALSE; for LSQ: TRUE).
- ORTHogonalize
- { [SELEction=<selection>]
[A=<real>] [B=<real>] [C=<real>]
[ALPHa=<real>] [BETA=<real>] [GAMMa=<real>] } orthogonalizes selected coordinates. The X-PLOR convention keeps the direction of the x-axis (x same direction as a; y is in (a,b) plane) and is the same that is used internally by all X-PLOR routines (default for selected atoms: (ALL); for a, b, c: 1.0; for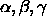 : 90
: 90 ).
).
- RGYRation
- { [SELEction=<selection>]
[MASS=<logical>]
[FACT=<real>] } computes radius of gyration
where the angle brackets denote averaging over selected atoms. The averaging is mass-weighted if MASS is TRUE. The factor FACT is subtracted from the masses before applying the mass-weighting. The symbols $RG (radius of gyration), $XCM, $YCM, $ZCM (center of mass) are declared (default for selected atoms: (ALL); for MASS: FALSE; for FACT: 0.0).
- RMS
- { [SELEction <selection>] [MASS=<logical>] } computes the (mass-weighted if MASS is TRUE) rms difference for selected atoms between the main and comparison set. The rms value is stored in the symbol $RESULT. The individual atomic rms differences are stored in the RMSD array (default for selection: (ALL); for MASS: FALSE).
- ROTAte
- { [SELEction=<selection>]
[CENTer=<3d-vector>]
<matrix> } rotates selected atoms around the specified rotation center (default: ( 0 0 0) ). The rotation matrix is specified through the matrix statement (see Section 2.4) (default for selection: (ALL)). - SHAKe
- { [MASS=<logical>] [REFErence=MAIN|COMP] } iteratively modifies main coordinate set until SHAKE constraints (see Section 8.2) are satisfied. The reference coordinate set specifies the direction of the SHAKE shift vectors (default for MASS: FALSE; for REFErence: MAIN).
- SWAP
- { [SELEction=<selection>] } exchanges main
and comparison
coordinate set, i.e.,
X,Y,Z
 XCOMP,YCOMP,ZCOMP; B,Q are unaffected
(default for selection: (ALL) ).
XCOMP,YCOMP,ZCOMP; B,Q are unaffected
(default for selection: (ALL) ).
- SYMMetry
- <symmetry-operator>
[SELEction=<selection>]
applies the specified
crystallographic symmetry operator
to the selected coordinates.
The notation for the symmetry operator
is the same as in the International Tables for Crystallography (Hahn ed. 1987),
e.g., (-x,
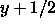 , -z). The unit cell
geometry needs to be specified (Section
12.3) before invoking this statement. The coordinates
are converted into fractional coordinates before application
of the symmetry operator and converted back into orthogonal
coordinates afterward.
, -z). The unit cell
geometry needs to be specified (Section
12.3) before invoking this statement. The coordinates
are converted into fractional coordinates before application
of the symmetry operator and converted back into orthogonal
coordinates afterward.
- TRANslate
- { [SELEction=<selection>]
VECTor=<3d-vector> [DISTance=<real>] }
translates selected atoms by specified translation
vector. If DISTance is specified, the translation occurs along
the specified vector for the specified distance (default for
selection: (ALL) ).
DERIvative | MAIN | REFErence ] [SELEction=<selection>]
{ <pdb-record> } reads Brookhaven Data Bank formatted records consisting of x,y,z coordinates, occupancies, and B-factors, tries to match the atom name, residue name, residue number, and segment name, and deposits the information in the main (X,Y,Z,B,Q), comparison (XCOMP, YCOMP, ZCOMP, BCOMP, QCOMP), reference (REFX, REFY, REFZ, HARM, HARM), or derivative (DX, DY, DZ, FBETA, FBETA) coordinate arrays. (For the definition of these arrays, see Section 2.16.) The information is deposited only if the atom has been selected. Note that the syntax is strict; i.e., the SELEction and the DISPosition have to be specified before one can read a <pdb-record>. Also, the PDB convention suggests an END statement at the end of the file that will terminate the coordinate statement (default for selected atoms: (ALL); for DISPosition: MAIN).