



Next: About this document
Up: X-PLOR On-line Manual
Previous: Abbreviations
, 
- ( )
- ( )
- 1--4 interaction
,  ,
,  ,
,  ,
,  ,
, 
- :==
- <* *>
, 
- < >
- R value
, 
- R value, distribution
- R value,free R value
- R value,Luzzati plot
- R value,NMR structure
, 
- |
- @
- @@
- accessible surface area
- anomalous scattering
- application
, 
- atom-based parameters
, 
- atomic form factors
- atom properties
- atom selection
- atom selection, fragile
- atom tag
, 
- B-factor refinement
, 
- Backus-Naur notation
- benchmark results
- bond angle deviations
- bond length deviations
- bound smoothing
- Brookhaven Protein Data Bank
- building,hydrogens
- carbohydrates,topologies and parameters
- centroid search
- CHARMM trajectory format
- chiral centers
- chirality
, 
- chromophores,topologies and parameters
- completeness of diffraction data
- conformational analysis
- conjugate gradient minimization
- constraints,coordinate
- constraints,fixing atomic positions
- constraints,fixing distances
- constraints,SHAKE
- constraints interaction statement
- control statement
- coordinates
- coordinates,best fit
- coordinates,comparison set
- coordinates, fractionalization
, 
- coordinates,manipulating
- coordinates, orthogonalization
, 
- coordinates,rms difference
- coordinates,rms difference (rmsd) analysis
- coordinates,rms difference analysis
- coordinates,rms difference,pairwise
- coordinates,rotation and translation
- coordinates, unknown
- coordinates,writing
- crystallographic diffraction data
- crystallographic refinement
- crystallographic refinement, atomic positions
- crystallographic refinement, simulated annealing
- crystallographic refinement, B-factors
, 
- crystallographic refinement,alternate conformations,disorder
- crystallographic refinement,bad contacts
- crystallographic refinement,initial structure check
- crystallographic refinement,linear correlation coefficient
- crystallographic refinement,occupancies
- crystallographic refinement,overview
- crystallographic refinement,phase accuracy
- crystallographic refinement,slow-cooling
- crystallographic refinement,special positions
- crystallographic refinement,target functions
- crystallographic refinement,topologies and parameters
- crystallographic refinement,weights
- crystal packing
- deletion of atoms
- deviation from ideality
- diffraction data
- diffraction data, completeness
- diffraction data, expanding
- diffraction data, reducing
- diffraction data,manipulation
- diffraction data,scaling
, 
- dihedral angle restraints
- dimensioning of X-PLOR
- display
- distance
geometry,van der Waals interactions
- distance,between atoms
- distance analysis
- distance difference matrix
- distance geometry
, 
- distance geometry,
accuracy parameter
- distance geometry,bond angle constraints
- distance geometry,bound smoothing
- distance geometry,dihedral angle constraints
- distance geometry,distance bounds
- distance geometry,embedded substructures
- distance geometry,embedding
- distance geometry,full-structure embedding
- distance geometry,improper
angle constraints
- distance geometry,metrization
- distance geometry,pseudoatoms
- distance geometry,regularization of coordinates
- distance geometry,rigid group
- distance geometry,SA-regularization of structures
- distance geometry,scaling
- distance geometry,triangle inequalities
- distance matrix
- distance restraints
- distance restraints, restraining functions
- distance restraints,averaging methods
- distance restraints,disulfide bonds
- distance restraints,hydrogen bonds
- distance restraints,reading
- distribution medium of X-PLOR
- disulfide bridge
- disulfide bridges
- duplicating the molecular structure
- electron density map
- electron density map,
 map
map
- electron density map,map file
- electron density map,omit map
- electron density map,SA-refined omit map
- embedding
- empirical energy function
- energy,analysis
- energy, bond
- energy, bond angle
- energy, conformational
- energy,crystallographic symmetry
- energy, dihedral angle
- energy, electrostatic
- energy, hydrogen bond
- energy, improper angle
- energy,interaction between selected atoms
- energy,intermolecular
, 
- energy,intramolecular
- energy,non-crystallographic symmetry
- energy, nonbonded
- energy,pick
- energy,print
- energy,second function
, 
- energy, torsion angle
- energy,turning on or off
- energy, van der Waals
- energy analysis, conformational terms
- energy analysis,nonbonded terms
- energy analysis,setting the weight
- energy evaluation
- energy function
, 
- energy minimization
- equal sign
- Eulerian angles
- evaluate
- example files,on-line
- fast Fourier transformation
,  ,
, 
- figure of merit
,  ,
, 
- filenames
- flags
- for ... in
- force-field
- fractional coordinates
- fractionalized coordinates
- fragile atom selection
- free R value
- Friedel mates
- geometric analysis
, 
- geometry,pick
- geometry,print
- harmonic restraints
- heavy-side function
- help on-line
- histidine protonation
- hydrogen-bond donors and acceptors
- hydrogen-bonding parameters
- hydrogen building
- hydrogens
, 
- hydrogens,naming convention
- if ... then
- inertia tensor
- installation of X-PLOR
- interchain interaction energy
- intramolecular interactions
- Langevin dynamics
, 
- Lattman's angles
- learning parameters
- Lee and Richards algorithm
- Lennard-Jones potential
, 
- letters,uppercase and lowercase
- lipids,topologies and parameters
- loops
,  ,
, 
- Luzzati plot
- manipulating coordinates
- manipulation,diffraction data
- Mathematica
- matrices
- Maxwellian distribution function
- messages
- metrix matrix distance geometry
- metrization
- minimization
- minimization,conjugate gradient
- minimization,rigid-body
- mirror images
- molecular dynamics
- molecular dynamics,angular distribution
- molecular dynamics,average displacement
- molecular dynamics,covariance
- molecular dynamics,density
- molecular dynamics,finite difference approximation
- molecular dynamics,merging files
- molecular dynamics,orienting trajectory frames
- molecular dynamics,pick properties
- molecular dynamics,power spectrum
- molecular dynamics,radial distribution function
- molecular dynamics,restarts
- molecular dynamics,simulation
,  ,
, 
- molecular dynamics,slow-cooling
- molecular dynamics,temperature control
- molecular dynamics,temperature control,Langevin dynamics
- molecular dynamics,temperature control,temperature coupling
- molecular dynamics,temperature control,velocity rescaling
- molecular dynamics,time correlation
- molecular dynamics,trajectories
- molecular dynamics,trajectory analysis
- molecular dynamics,velocity assignment
- molecular replacement
- molecular replacement,cluster analysis
- molecular replacement,cluster criteria
- molecular replacement,comparing rotation matrices
- molecular replacement,contour plots
- molecular replacement,cross rotation function,PC-refinement
, 
- molecular replacement,cross-rotation function
- molecular replacement,direct rotation function
- molecular replacement,elbow angle modification
- molecular replacement,packing function
- molecular replacement,rigid-body refinement
- molecular replacement,rotation search
- molecular replacement,self-rotation function
- molecular replacement,translation function,asymmetric unit
- molecular replacement,translation function,finding second molecule
- molecular replacement,translation function,relative z position
- molecular replacement,translation function,using PC-refined model
- molecular replacement,translation search
- molecular structure file
- molecular structure,append
- molecular structure,deletion
- molecular structure,disulfide bridge
- molecular structure,duplicating
- molecular structure,examples
- molecular structure,generation
- molecular structure,histidine protonation
- molecular structure,incomplete polypeptide chain
- molecular structure,nucleic acid
- molecular structure,patching
, 
- molecular structure,polypeptide chain
- molecular structure,protein
- molecular structure,protein,unusual geometries
- molecular structure,reading
- molecular structure,sequence
- molecular structure,solvent and solute
- molecular structure,unknown atoms
- molecular structure,viruses or multimers
- molecular structure,water
- molecular structure,writing
- NCS
, 
- Newton's equations of motion
- NMR, relaxation matrix refinement
- NMR relaxation refinement, R value
- NMR relaxation refinement,against NOESY intensities
- NMR relaxation refinement,cutoffs
- NMR relaxation refinement,including solvent spectra
- NMR relaxation refinement,input of experimental data
- NMR relaxation refinement,optimal correlation time
- NMR relaxation refinement,quality assessment
- NMR structure determination
- NMR structure determination, topologies and parameters
- NMR structure determination,acceptance of refined structures
- NMR structure determination,average structure
- NMR structure determination,enantiomer selection
- NMR structure determination,rms differences
, 
- NMR structure determination,simulated annealing
,  ,
, 
- NMR structure determination,substructure distance geometry
- NMR structure determination,template coordinate set
- NMR structure refinement, topologies and parameters
- NOE
- NOE, 3D
- NOE,time-average
- NOE back-calculation
- NOE back-calculation,BPTI example
- NOE back-calculation,gradient expression
- NOE back-calculation,leakage
- NOE back-calculation,unresolved chemical shifts
- NOE distances, restraints
- NOE distances,averaging methods
- NOE distances,distance restraints file
- NOE distances,pseudoatoms
- NOE distances,restraining functions
- non-crystallographic symmetry
- non-crystallographic symmetry,Bricogne conventions
- non-crystallographic symmetry,restrained
- non-crystallographic symmetry,restraints
- non-crystallographic symmetry,skew frame
- non-crystallographic symmetry,strictly identical
, 
- nonbonded distances
- nonbonded energy terms
, 
- nonbonded energy terms,truncation
- nonbonded interaction, 1--4
,  ,
,  ,
,  ,
,  ,
, 
- nonbonded parameters
- nucleic acids,molecular structure
- nucleic acids,topologies and parameters
,  ,
,  ,
, 
- numerical precision
- occupancy refinement
- orthogonalized coordinates
, 
- packing function
, 
- parameter,writing
- parameter and topology files
- parameter and topology files, lipids
- parameter and topology files, water
- parameter and topology files,carbohydrates
- parameter and topology files,chromophores
- parameter and topology files,crystallographic refinement
- parameter and topology files,nucleic acids
,  ,
,  ,
, 
- parameter and topology files,proteins
,  ,
,  ,
,  ,
, 
- parameters
- parameters, atom-based
, 
- parameters, hydrogen-bonding
- parameters,learning
- parameters, nonbonded
- parameters,reducing
- parameters, type-based
- parsing
- partial energy terms
- PATCh statement
- Patterson correlation (PC) refinement
- PDB format
- phase difference
- phase restraints
- pick energy
- pick geometry
- planarity
- Powell minimization
- print energy
- print geometry
- proteins,molecular structure
- proteins,topologies and parameters
,  ,
,  ,
,  ,
, 
- pseudoatoms
,  ,
, 
- quanta
- QUANTA trajectory format
- quaternions
, 
- radius of gyration
, 
- Ramachandran plot
- random-number function
- random-number seed
- reading,coordinates
- reading,distance restraints
- reading,molecular structure
- reading,trajectory
- reducing parameters
- reflection files
, 
- reflection files,merging files
- reflections,manipulation
- reflections,writing
- relaxation energy term
- relaxation matrix
- relaxation matrix refinement, NMR
- remarks
- repel energy term
- residues
- restraints,coordinate
- restraints,dihedral
- restraints, dihedral angle
- restraints,distance
- restraints,distance symmetry
- restraints, harmonic plane
- restraints, harmonic point
- restraints,harmonic coordinate
- restraints,planarity
- rigid-body minimization
- rigid-body molecular dynamics
- rigid-body refinement
- ring pucker
, 
- ring pucker,nucleic acids
- rmsd
,  ,
, 
- rms deviation
- rotation function,list file
- rotation function,output file
- rotation matrix (3
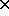 3)
3)
- rotation search
- scaling of diffraction data
, 
- segment statement
- selection
- SHAKE
- simulated annealing, ab initio from template
- simulated annealing,crystallographic refinement
- simulated annealing,NMR refinement
- simulated annealing,preparation
- simulated annealing,random coordinates
- simulated annealing,slow-cooling
,  ,
, 
- slow-cooling,crystallographic refinement
- slow-cooling molecular dynamics
- solvation
- solvent mask
, 
- solvent screening
- special positions
- structure,generation
- structure factor,normalized
- structure factor,partial
, 
- structure factor,solvent
- structure factors
- structure generation,nucleic acid
- structure generation,protein
- structure generation,protein,with cofactor or substrate
- structure generation,protein,with metal cluster
- structure generation,protein,with water or ligands
- structure generation,viruses or multimers
- surface statement
- symbols
- symmetry,distance restraints
- symmetry interactions,crystallographic
- symmetry interactions,non-crystallographic
- symmetry operators
,  ,
,  ,
, 
- tag
, 
- time-averaged distance restraints
- topology
- trajectory,analysis
- trajectory,analysis,angular distribution
- trajectory,analysis,average
- trajectory,analysis,covariance
- trajectory,analysis,density
- trajectory,analysis,pick properties
- trajectory,analysis,power spectrum
- trajectory,analysis,radial distribution function
- trajectory,analysis,time correlation
- trajectory,error messages
- trajectory,merging coordinate files
- trajectory,merging files
- trajectory,reading
- trajectory,writing
- trajectory statement
- translation function,output file
- translation function,peak list
- translation search
- type-based parameters
- unit cell constants
- unknown coordinates
- Van der Waals radii, parameters
- van der Waals radius
- van der Waals radius, in NMR protocols
- vector
- vectors (3d)
- Verlet method
- viruses,molecular structure
- water
, 
- water,topologies and parameters
- water hydrogens
- while
- wildcards
- Wilson plot
, 
- writing,coordinates
- writing,molecular structure
- writing,reflections
- writing,trajectory
- X-PLOR,dimensioning
- X-PLOR,input
- X-PLOR,installation
- X-PLOR,list of subdirectories
- X-PLOR,notation
Web Manager
Sat Mar 11 09:37:37 PST 1995
 map
map 3)
3) map
map 3)
3)