




Next: Write Coordinate Statement
Up: Coordinate Statement
Previous: Requirements
The first example reads the coordinates from two files into the main
and comparison sets:
coordinates @set1.pdb
coordinates disposition=comparison @set2.pdb
In the second example, the first set is least-squares fitted to the second
(comparison) set using 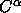 atoms only. Note that all atoms
are translated and rotated. Then the rms difference
between the two sets is computed for backbone atoms and stored
in the symbol $1. Finally, the individual rms differences
are printed for all backbone atoms that show rms differences
greater than 1 Å.
atoms only. Note that all atoms
are translated and rotated. Then the rms difference
between the two sets is computed for backbone atoms and stored
in the symbol $1. Finally, the individual rms differences
are printed for all backbone atoms that show rms differences
greater than 1 Å.
coordinates fit selection=( name ca ) end
coordinates rms selection=( name ca or name n or name c ) end
evaluate ($1=$result)
vector show ( rmsd )
( attribute rmsd > 1.0 and ( name ca or name n or name c ))
In the third example, the main coordinates are fractionalized,
a translation and a rotation are applied, and the coordinates
are orthogonalized again. The crystallographer will recognize the
rotation and translation as the space-group operator 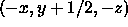 .
.
coordinates fractionalize
a=30. b=40. c=20. alpha=90. beta=100. gamma=90.
end
coordinates rotate
matrix=( -1 0 0 )
( 0 1 0 )
( 0 0 -1 )
end
coordinates translate
vector=( 0 0.5 0 )
end
coordinates orthogonalize
a=30. b=40. c=20. alpha=90. beta=100. gamma=90.
end
The final example shows how to rotate the coordinates
using Eulerian angles:
coordinates rotate
euler=( 10., 30., 2. )
end
Web Manager
Sat Mar 11 09:37:37 PST 1995
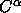 atoms only. Note that all atoms
are translated and rotated. Then the rms difference
between the two sets is computed for backbone atoms and stored
in the symbol $1. Finally, the individual rms differences
are printed for all backbone atoms that show rms differences
greater than 1 Å.
atoms only. Note that all atoms
are translated and rotated. Then the rms difference
between the two sets is computed for backbone atoms and stored
in the symbol $1. Finally, the individual rms differences
are printed for all backbone atoms that show rms differences
greater than 1 Å.
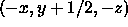 .
.