




Next: Requirements
Up: Rotation Search
Previous: Rotation Search
- SEARch Rotation
-
{ <xrefin-search-rotation-statement> } END
is an xrefin statement. Action takes place as soon as the END
statement is issued.
- <xrefin-search-rotation-statement>
- :==
- DELTa=<real>
- specifies an angular grid interval.
DELTa should be less than
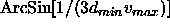 ,
where
,
where  is the high-resolution limit (see Section
12.3) and
is the high-resolution limit (see Section
12.3) and 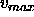 is the maximum Patterson vector length
of the search model (see Section 17.5.3 for an example).
This is a conservative estimate. In many cases, it will
be sufficient to double this estimate. Note that the CPU time
requirement for the rotation function is roughly an inverse
cubic function of DELTa. Thus, one should use the largest
possible value for DELTa (default: 0.0).
is the maximum Patterson vector length
of the search model (see Section 17.5.3 for an example).
This is a conservative estimate. In many cases, it will
be sufficient to double this estimate. Note that the CPU time
requirement for the rotation function is roughly an inverse
cubic function of DELTa. Thus, one should use the largest
possible value for DELTa (default: 0.0).
- EPSIlon=<real>
- specifies
 for
the peak cluster analysis. Multiple entries
produce multiple cluster analysis (default: 0.2).
for
the peak cluster analysis. Multiple entries
produce multiple cluster analysis (default: 0.2).
- FORMatted=<logical>
- should be set to FALSE if
the map files are binary files (default: TRUE).
- LIST=<filename>
- specifies the
file for the list of rotation function
peaks after the cluster analysis (grid points are mapped into
an asymmetric unit of the rotation function) (default: OUTPUT).
- NLISt=<integer>
- specifies that the analysis of the
rotation function will be restricted to
the <integer> highest grid points.
- NPEAks=<integer>
- selects the <integer> strongest
Patterson vectors of map P1 that satisfy the
range and threshold criteria (default: 5000).
- OUTPut=<filename>
- specifies the file for the complete rotation function.
- P1INput=<filename>
- reads Patterson map P1.
- P2INput=<filename>
- reads Patterson map P2.
- PSIMIn=<real>
- PSIMAx=<real> PHIMIn=<real>
PHIMAx=<real> KAPPAMIn=<real>
KAPPAMAx=<real>
is spherical polar angle sampling (as defined in Section
2.4). Specified are the
minimum and maximum values for 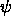 ,
, 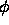 ,
,  (default: 0).
(default: 0).
- RANGe=<real>=<real>
- selects the Patterson
vectors of map P1 based on
the distance of the vectors from the origin
(default: 3--20 Å).
- SELF=<logical>
- is a flag for cluster analysis that
determines the symmetry of map P1. If FALSE, map P1 is
treated with
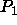 symmetry, whereas map P2 is treated with
crystallographic symmetry, as in
the xrefin symmetry statement. If TRUE, both P1
and P2 are treated with crystallographic symmetry.
symmetry, whereas map P2 is treated with
crystallographic symmetry, as in
the xrefin symmetry statement. If TRUE, both P1
and P2 are treated with crystallographic symmetry.
- T1MIn=<real>
- T1MAx=<real> T2MIn=<real>
T2MAx=<real> T3MIn=<real> T3MAx=<real>
is equidistant Eulerian angle sampling
(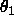 ,
, 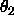 ,
,  , as defined in
Section
2.4) using DELTa intervals.
Specified are the minimum and maximum values for
, as defined in
Section
2.4) using DELTa intervals.
Specified are the minimum and maximum values for
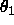 ,
, 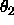 ,
, 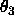 (default: 0 ).
(default: 0 ).
- THREshold=<real>
- selects the Patterson vectors of map P1
according to the value of the Patterson map: if
this value is larger than the specified threshold,
the Patterson vector is selected (default: 500).
- TPMIn=<real>
- TPMAx=<real>
TMMIn=<real>
TMMAx=<real> T2MIn=<real>
T2MAx=<real>
is Lattman's (1972) pseudo-orthogonal sampling, using
DELTa as the interval for 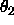 . Specified are the
minimum and maximum values for
. Specified are the
minimum and maximum values for 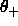 ,
, 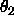 ,
and
,
and 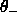 (default: 0 and none).
(default: 0 and none).





Next: Requirements
Up: Rotation Search
Previous: Rotation Search
Web Manager
Sat Mar 11 09:37:37 PST 1995
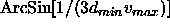 ,
where
,
where  is the high-resolution limit (see Section
12.3) and
is the high-resolution limit (see Section
12.3) and 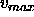 is the maximum Patterson vector length
of the search model (see Section 17.5.3 for an example).
This is a conservative estimate. In many cases, it will
be sufficient to double this estimate. Note that the CPU time
requirement for the rotation function is roughly an inverse
cubic function of DELTa. Thus, one should use the largest
possible value for DELTa (default: 0.0).
is the maximum Patterson vector length
of the search model (see Section 17.5.3 for an example).
This is a conservative estimate. In many cases, it will
be sufficient to double this estimate. Note that the CPU time
requirement for the rotation function is roughly an inverse
cubic function of DELTa. Thus, one should use the largest
possible value for DELTa (default: 0.0).
 for
the peak cluster analysis. Multiple entries
produce multiple cluster analysis (default: 0.2).
for
the peak cluster analysis. Multiple entries
produce multiple cluster analysis (default: 0.2).
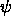 ,
, 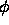 ,
,  (default: 0).
(default: 0).
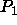 symmetry, whereas map P2 is treated with
crystallographic symmetry, as in
the xrefin symmetry statement. If TRUE, both P1
and P2 are treated with crystallographic symmetry.
symmetry, whereas map P2 is treated with
crystallographic symmetry, as in
the xrefin symmetry statement. If TRUE, both P1
and P2 are treated with crystallographic symmetry.
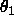 ,
, 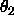 ,
,  , as defined in
Section
2.4) using DELTa intervals.
Specified are the minimum and maximum values for
, as defined in
Section
2.4) using DELTa intervals.
Specified are the minimum and maximum values for
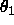 ,
, 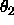 ,
, 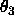 (default: 0 ).
(default: 0 ).
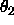 . Specified are the
minimum and maximum values for
. Specified are the
minimum and maximum values for 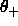 ,
, 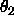 ,
and
,
and 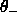 (default: 0 and none).
(default: 0 and none).