The symmetry function restrains distance differences between pairs of ASSIgn statements. The distance pairs should be entered as follows:
assign (segid a and resid 1 and name ca) {* First distance.*}
(segid b and resid 10 and name ca) 0.0 0.1 0.1
{* Second distance.*}
assign (segid a and resid 10 and name ca)
(segid b and resid 1 and name ca) 0.0 0.0 0.0
This particular example restrains the signed
difference between the
first and second distance to 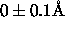 .
The values of r,
.
The values of r, 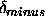 , and
, and 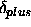 for the
second distance are not used by the program.
for the
second distance are not used by the program.
The list of symmetry-related intermonomer distance differences for a dimeric protein can be generated automatically by the following script, which produces a file ``symrest.tbl":
{* Name of the distance restraints file.*}
set display=symrest.tbl end
{* Get number of residues in monomer.*}
vector do (store1 = decode(resid)) (segid a and name ca)
vector show min (store1) (segid a and name ca)
evaluate ($first_residue = $result)
vector show max (store1) (segid a and name ca)
evaluate ($last_residue = $result)
for $id in id (name ca and segid a) loop res1
vector show element (resid) (id $id)
evaluate ($resid1 = decode($result))
evaluate ($resid2 = $last_residue - $resid1 + $first_residue)
if ($resid2 > $resid1) then
display ! distance pair $resid1 $resid2
display assign (resid $resid1 and name ca and segid a)
display (resid $resid2 and name ca and segid b) 0 0 0
display assign (resid $resid1 and name ca and segid b)
display (resid $resid2 and name ca and segid a) 0 0 0
end if
end loop res1
The resulting distance difference list will restrain the
distance differences to 0; i.e., it will attempt to produce
a perfectly symmetric arrangement of the dimer.
The next example shows how one would actually use the symmetry restraints:
noe class symm @symmetry.tbl potential symm symmetry scale symm 1.0 sqconstant symm 1.0 sqexponent symm 2 soexponent symm 1 rswitch symm 0.5 sqoffset symm 0.0 asymptote symm 1.0 endNote that these symmetry distance restraints are more stringent than the non-crystallographic symmetry restraints described in Section 16.1. The distance symmetry implies twofold symmetry whereas the non-crystallographic symmetry restraints simply restrain the monomers to be nearly superimposable without specification of a specific operation between the monomers. The distance symmetry restraints are, however, less stringent than the strict non-crystallographic symmetry (Section 16.2), as the latter requires explicit specification of the symmetry operation. The distance symmetry restraints allow the separation between the monomers to be a self-adjusting parameter. (For further reading, see Nilges and Brünger 1991.)