




Next: NMR Structure Determination
Up: Distance Geometry
Previous: Regularization
The X-PLOR/DG
code is highly vectorizable and parallelizable and should
perform efficiently on most modern computer architectures.
The complexity of full metrization is 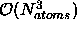 ,
where
,
where 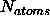 is the number of atoms in the (sub) structure. The
complexity of bound smoothing alone depends on the number
of atoms and on the number of connectivities (known distances) in the
molecule.
For a molecule consisting of 500 atoms, it takes about 300s
on a CONVEX 210 to
embed the molecule using partial (4-atom) random metrization.
is the number of atoms in the (sub) structure. The
complexity of bound smoothing alone depends on the number
of atoms and on the number of connectivities (known distances) in the
molecule.
For a molecule consisting of 500 atoms, it takes about 300s
on a CONVEX 210 to
embed the molecule using partial (4-atom) random metrization.
The X-PLOR/DG module allocates about
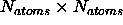 single
precision words of memory. (Section 20.11 contains more
CPU timing results.)
single
precision words of memory. (Section 20.11 contains more
CPU timing results.)
Web Manager
Sat Mar 11 09:37:37 PST 1995
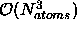 ,
where
,
where 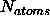 is the number of atoms in the (sub) structure. The
complexity of bound smoothing alone depends on the number
of atoms and on the number of connectivities (known distances) in the
molecule.
For a molecule consisting of 500 atoms, it takes about 300s
on a CONVEX 210 to
embed the molecule using partial (4-atom) random metrization.
is the number of atoms in the (sub) structure. The
complexity of bound smoothing alone depends on the number
of atoms and on the number of connectivities (known distances) in the
molecule.
For a molecule consisting of 500 atoms, it takes about 300s
on a CONVEX 210 to
embed the molecule using partial (4-atom) random metrization.
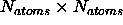 single
precision words of memory. (Section
single
precision words of memory. (Section