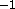 for the final step of minimization.
This protocol has been successfully used with up to three disulfide
bridges. However, in the case of bovine pancreatic trypsin
inhibitor, removal of the disulfides was required.
for the final step of minimization.
This protocol has been successfully used with up to three disulfide
bridges. However, in the case of bovine pancreatic trypsin
inhibitor, removal of the disulfides was required.
The next step involves generation of a template coordinate set. This is required in all cases, except for the random simulated annealing protocol. The template coordinate set can be any conformation of the macromolecule with good local geometry and no nonbonded contacts. It can be generated by using most molecular modeling graphics programs or, preferably, by using the X-PLOR protocol described below. The purpose of the template coordinate set is to provide distance geometry information about the local geometry of the macromolecule, to apply appropriate pseudoatom corrections, and to provide a starting point for the ab initio simulated annealing protocol. Furthermore, template coordinates can be used to define large rigid groups in distance geometry, e.g., if certain portions of the protein structure are known by other means or have been determined by other methods.
The protocol below automatically generates a template coordinate set. It initially places the atoms of the macromolecule along the x-axis, with y and z set to random numbers. The coordinates are then regularized using simulated annealing. The protocol is completely general, and it has been tested for both proteins and nucleic acids.
Disulfide bonds and other covalent links between
sequentially distant residues may have to be
removed for successful completion of the template
generation. Generally, when too many covalent
links are present, the structure may get entangled
in a knot which will result in poor local geometry. Some experimentation
may be required to find out if certain covalent links
have to be removed; the goal is to obtain an energy
below 1000 kcal mole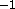 for the final step of minimization.
This protocol has been successfully used with up to three disulfide
bridges. However, in the case of bovine pancreatic trypsin
inhibitor, removal of the disulfides was required.
for the final step of minimization.
This protocol has been successfully used with up to three disulfide
bridges. However, in the case of bovine pancreatic trypsin
inhibitor, removal of the disulfides was required.