




Next: Requirements
Up: Setup of the
Previous: Setup of the
- RELAxation { <relaxation-statement> } END
- is
invoked from the
main level of X-PLOR.
- <relaxation-statement>:==
-
- ASSIgn
- <selection> <selection> {
 }
adds a single cross-peak intensity or a
buildup curve between the two
specified selected sets of spins. The number of mixing times
read depends on the number of CLASs statements issued before the
ASSIgn statement.
Error estimates can be specified. The
format depends on the ERROr_input statement. For
examples, see Section 21.7.
}
adds a single cross-peak intensity or a
buildup curve between the two
specified selected sets of spins. The number of mixing times
read depends on the number of CLASs statements issued before the
ASSIgn statement.
Error estimates can be specified. The
format depends on the ERROr_input statement. For
examples, see Section 21.7.
- AVERage <real>
- specifies an exponent for calculating the
average distance to an UNREsolved group of protons (default: 6).
- CALIbrate { <calibrate-statement> } END
- calculates the calibration
factor between the observed and calculated intensities. Prior to the
calibration, this command computes the NOE intensities from the current
atomic model unless VALUe is specified.
- <calibrate-statement>:==
-
- AUTOmatic=<ON,OFF>
- sets the calibration factor, if ON,
every step during the refinement. At present, this allows the calculation
of the gradient only approximately. Therefore, it should be turned OFF during
minimization.
- GROUp=<CLASs,GROUp,ALL>
- for CLASS, calculates the
calibration factor
separately for each classification; for ALL, one overall
calibration factor is used (default: CLASs).
- QUALity=<real>
- specifies that cross peaks for which the
relative error estimate exceeds the specified value are not used (default: 1).
- REFErence=<ALL,REFErence>
- for ALL, uses all peaks that
meet the criterion set with the EXCLude option for the
calculation of the calibration factor; for REFErence, uses only those
that were flagged as reference peaks during input (default: ALL).
- VALUe class <*real*>
- sets a value for the calibration factor.
Note: if the calibration factor is set by VALUe for
a class, it is not recalculated for other classes
in the same CALIbrate statement.
- CLASsification <class-name>
- partitions an intensity
table; it applies to all following ASSIgn entries
until another CLASS entry is issued.
In contrast to the NOE CLASs statement, the RELAxation CLASs
statement can be used to switch between classes during the input
of cross-peak intensities. Several class statements can be
issued without an intervening ASSIgn statement. This allows
the input of buildup curves (default: NIL).
- CLGR (CLass_GRoup) <*class*> <group>
-
enters a CLASs into a GROUp of spectra (default: none).
- CUTOff
- { <cutoff-statement> } END applies a cutoff to
relaxation matrix and/or gradient calculation. When a cutoff is in
effect, for each peak
on the data list a separate small relaxation matrix is set up,
including only spins
within a range of the spin pair specified by the cutoff.
The spectrum and the gradient are calculated with this
small relaxation matrix.
- <cutoff-statement>:==
-
- MODE=<NONE,GRADient,ALL>
- applies no cutoff, cutoff only
for the
gradient calculation, or cutoff for both intensity and gradient
calculation
(default: NONE).
- RELAtive_value=<real>
- specifies a cutoff relative to the actual distance
between the two spins in Å (default: 0).
- VALUe=<real>
- specifies the actual value of the cutoff,
in Å (default: 6).
- EEXPonent <real>
- is an exponent m of the WELL or
PARAbolic energy function (default: 2).
- ERROr_input
- { <error-input-statement> } END
defines an error input format for the following ASSIgn statement.
It remains in effect until another ERROr_input command is
issued.
- <errorinput-statement>:==
-
- INPUt=<UNIForm,RANGe,PLUSminus>
- specifies the
input format (0, 1, or 2
error estimates) for the following ASSIgn statements
(default: RANGe): for UNIForm, no error estimate is expected
by the program, for RANGe, one error estimate, and for PLUSminus,
two, where the first is an error estimate on the minus side, the second on the plus side.
- MODE=<ABSOlute,RELAtive>
- interprets the error estimate in the
ASSIgn statements as absolute or relative
(default: ABSOlute).
- VALUe=<real>
- enters the actual value for UNIForm option
(default: 0).
- GROUp <group-name>
- defines a group of several
CLASses that are recorded with the same physical conditions,
like solvent(H
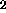 O, D
O, D O), field strength, and temperature
(default: NIL).
O), field strength, and temperature
(default: NIL).
- IEXPonent <real>
- specifies an exponent n of the calculated and
observed intensities in the energy function (default: 1).
- IWEIght
- { <iweight-statement> } END applies
individual weights
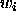 to each single term in the energy function.
At the moment, the only supported options are weights equal to the inverse
of
powers of the observed intensity (i.e.,
to each single term in the energy function.
At the moment, the only supported options are weights equal to the inverse
of
powers of the observed intensity (i.e.,  ).
If the exponent g matches the exponent m of the energy
function, the weighting is equivalent to refining relative intensity
differences. The minimum weight applied is always 1.
).
If the exponent g matches the exponent m of the energy
function, the weighting is equivalent to refining relative intensity
differences. The minimum weight applied is always 1.
- <iweight-statement>:==
-
- CUTOff=<real>
- is the maximum weight applied (default: 10000).
- EXPOnent=<real>
- specifies an exponent g of the
observed intensity in the weight function
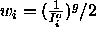 (default: 2).
(default: 2).
- QUALity=<real>
- stipulates that cross peaks for which the
relative error estimate exceeds the specified value are not used in
determining the maximum weight (default: 1).
- MINIntensity <*class*> <real>
- sets the minimum
detected intensity for each class.
The specified value is used as the ``zero" for the intensities (default:
smallest number used by X-PLOR).
- NREStraints <integer>
-
is the required parameter that specifies
the maximum expected number of intensities.
It has to be greater than or equal to the actual number. The parameter is
used to
assign space dynamically for the intensity list (default: 200).
- OCCUpancy <group> <selection> <real>
- sets
the occupancy for selected spins (for example, 0.9 for exchanging
amid protons) (default: 1.0).
- OMEGa <*group*> <real>
- specifies the spectrometer frequency, in Hz
(default: 500*10
 ).
).
- OMIT <group> <selection>
- is an obsolete statement
that was used
in a prerelease version of the relaxation matrix code of X-PLOR. It will
be removed in a future version. The statement deletes spins from the
relaxation matrix. This is almost the reverse of the SELEct
command, but it leaves any existing base of unresolved groups
intact (default: none).
- POTEntial <WELL,PARAbolic>
- provides two alternatives: WELL uses a flat-bottom
potential with bounds at error estimates, PARAbolic a
(generalized) parabola with exponent EEXP and center at
the specified intensity (default: WELL).
- PREDict_intensities
- <class>
{ <predict-intensities-statement> } END
calculates a spectrum from coordinates with the parameters valid for the
specified class. Note that this destroys any existing experimental intensity database.
The spectrum can be calculated for parts of a molecular structure.
- <PREDict_intensities-statement>:==
-
- CUTOFf <real>
- specifies a distance cutoff for cross-peak
calculation (default: 10).
- CUTON <real>
- specifies a distance cuton for cross-peak
calculation (default: 0).
- DIAGonal <ON,OFF>
- prints diagonal peaks (default: OFF).
- FORMat <NORMal,LIST>
- is an output format; LIST produces
an intensity INPUT file (default: NORMal).
- FROM <selection>
- selects the first part of a molecular structure
(default: all NMR-active atoms).
- THREshold <real>
- specifies the minimum intensity that is printed (default: 0).
- TO <selection>
- selects the second part of a molecular structure
(default: all NMR-active atoms).
- PRINt_deviations
-
{ <print-deviation-statement> } END
prints the deviations from observed intensities and calculates
an R value. It uses the form of the energy function
( WELL or PARAbolic and the value of IEXP) specified
before issuing the statement.
- <print-deviation-statement>:==
-
- FORMat <NORMal,LIST>
- is an output format; LIST produces
a listing in the format of an intensity input file.
- SELEct <selection>
- calculates the R value with only a
subset of the atoms. This allows the calculation of
local R values.
- THREshold <real>
- specifies the minimum deviation that is printed,
in the arbitrary units of the input intensities.
For a complete list of deviations (including 0), THREshold
should be set to a negative number (default: 0).
- REFErence
- flags intensities read by the
following ASSIgn
statements as reference peaks for the calculation of the calibration
factor (see CALIbrate statement),
and remains in effect until another CLASs statement is issued.
This flag allows one to use a subset of peaks for the calibration,
e.g., peaks that do not depend very much on the conformation
of the molecule.
The reference peaks are written into a separate file,
and the reference flag is set before reading that file.
- RESEt
- erases the current relaxation database.
- SELEct <group> <selection>
- selects the ``NMR-active"
atoms (default: (HYDRogen) ).
At present, only protons can be used in the refinement. The
NMR parameters for protons are applied to all selected atoms.
This command initializes the array that stores information about
unresolved groups (see below).
If the command is not issued at the beginning of the relaxation matrix setup,
all hydrogens are used by default. Since a hydrogen is defined by
its mass in X-PLOR, care should be taken in this case to modify the
masses of hydrogens for simulated
annealing only after the relaxation matrix parameters
have been set up.
- SORT_intensities
- sorts the
intensity list so that peaks with different mixing times
belonging to the same spin pair are consecutive on the list.
If a cutoff is used, a new relaxation matrix generally has to be set up and
diagonalized for each peak on the data list. However,
if the only difference between two entries on the data list is the mixing time,
the relaxation matrix does not have
to be recalculated and diagonalized. Sorting can thus save a considerable
amount of CPU time.
- TAUCorrelation
- <group> { <TAUC-statement> } END
specifies the correlation time and the order parameter, depending
on the chosen model.
- <TAUCorrelation-statement>:==
-
- MODEl <RIGId|LIPAri>
- provides two alternatives: RIGId
expects no order parameter;
LIPAri expects one overall correlation time and one order parameter.
This applies to both the ISOTropic and the VECTor case.
- ISOTropic
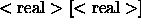
- specifies an
isotropic correlation time
and an order parameter (default: 10 ns, 1).
- RESEt
- initializes a table.
- VECTor <selection> <selection>

- specifies a
correlation time
(in seconds) and an order
parameter for a specified vector.
- TAUMix <*class*> <real>
- specifies the mixing time
 , in seconds (default: 0.1 sec).
, in seconds (default: 0.1 sec).
- TOLErance <real>
- if not equal to zero,
approximates the exact calculation of the
 energy function and its first
derivatives with respect to the atomic coordinates
by freezing
energy function and its first
derivatives with respect to the atomic coordinates
by freezing  until atoms
have moved by less than TOLErance Å\
from the point at which
until atoms
have moved by less than TOLErance Å\
from the point at which  was
last computed. This is particularly useful for
molecular dynamics calculations. It should be set to 0
during minimization to avoid inconsistencies
in the force field (default: 0).
was
last computed. This is particularly useful for
molecular dynamics calculations. It should be set to 0
during minimization to avoid inconsistencies
in the force field (default: 0).
- UPDate
- computes the NOE intensities from the current atomic model.
- UNREsolved
-
<group> { <unresolved-statement> } END
replaces protons with unresolved chemical shifts by a single
spin. The distance to an unresolved group is calculated
as
 , where the exponent av is set with the
AVERage command. A diagonal leakage rate is added
to the relaxation matrix for each unresolved group.
The main use of this command is for methyl groups.
, where the exponent av is set with the
AVERage command. A diagonal leakage rate is added
to the relaxation matrix for each unresolved group.
The main use of this command is for methyl groups.
- <unresolved-statement>:==
-
- 1-2
- treats each methylene group as an unresolved group of
protons. A methylene group is recognized by
X-PLOR as two hydrogens bound to a heavy
atom.
- METHyl
- treats each methyl group as an unresolved group of
protons. A methyl group is recognized by
X-PLOR as three hydrogens bound to a
heavy atom.
- NONE
- deletes all previously defined unresolved groups.
- SELECT <selection>
- treats the selected atoms as
an unresolved group of protons.
- WEIGht <*class*> <real>
- specifies the
energy constant
 (overall weight)
(default: 1).
(overall weight)
(default: 1).
- ZLEAkage_rate <*group*>
- specifies the overall diagonal
leakage rate
 , in sec
, in sec (default: 0).
(default: 0).





Next: Requirements
Up: Setup of the
Previous: Setup of the
Web Manager
Sat Mar 11 09:37:37 PST 1995
 }
adds a single cross-peak intensity or a
buildup curve between the two
specified selected sets of spins. The number of mixing times
read depends on the number of CLASs statements issued before the
ASSIgn statement.
Error estimates can be specified. The
format depends on the ERROr_input statement. For
examples, see Section 21.7.
}
adds a single cross-peak intensity or a
buildup curve between the two
specified selected sets of spins. The number of mixing times
read depends on the number of CLASs statements issued before the
ASSIgn statement.
Error estimates can be specified. The
format depends on the ERROr_input statement. For
examples, see Section 21.7.
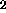 O, D
O, D O), field strength, and temperature
(default: NIL).
O), field strength, and temperature
(default: NIL).
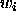 to each single term in the energy function.
At the moment, the only supported options are weights equal to the inverse
of
powers of the observed intensity (i.e.,
to each single term in the energy function.
At the moment, the only supported options are weights equal to the inverse
of
powers of the observed intensity (i.e.,  ).
If the exponent g matches the exponent m of the energy
function, the weighting is equivalent to refining relative intensity
differences. The minimum weight applied is always 1.
).
If the exponent g matches the exponent m of the energy
function, the weighting is equivalent to refining relative intensity
differences. The minimum weight applied is always 1.
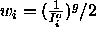 (default: 2).
(default: 2).
 ).
).
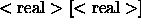

 , in seconds (default: 0.1 sec).
, in seconds (default: 0.1 sec).
 energy function and its first
derivatives with respect to the atomic coordinates
by freezing
energy function and its first
derivatives with respect to the atomic coordinates
by freezing  until atoms
have moved by less than TOLErance Å\
from the point at which
until atoms
have moved by less than TOLErance Å\
from the point at which  was
last computed. This is particularly useful for
molecular dynamics calculations. It should be set to 0
during minimization to avoid inconsistencies
in the force field (default: 0).
was
last computed. This is particularly useful for
molecular dynamics calculations. It should be set to 0
during minimization to avoid inconsistencies
in the force field (default: 0).
 , where the exponent av is set with the
AVERage command. A diagonal leakage rate is added
to the relaxation matrix for each unresolved group.
The main use of this command is for methyl groups.
, where the exponent av is set with the
AVERage command. A diagonal leakage rate is added
to the relaxation matrix for each unresolved group.
The main use of this command is for methyl groups.
 (overall weight)
(default: 1).
(overall weight)
(default: 1).
 , in sec
, in sec (default: 0).
(default: 0).