#include <coor.h>
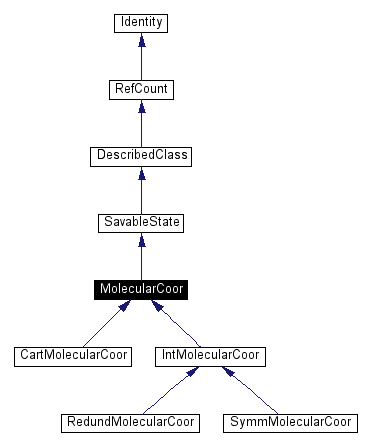
Inheritance diagram for MolecularCoor

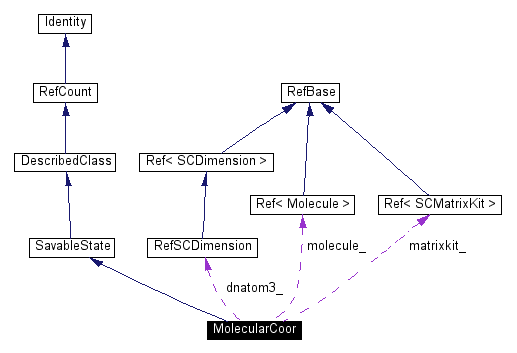
Public Methods | |
| MolecularCoor (Ref< Molecule > &) | |
| MolecularCoor (StateIn &) | |
| MolecularCoor (const Ref< KeyVal > &) | |
| The KeyVal constructor. More... | |
| virtual | ~MolecularCoor () |
| void | save_data_state (StateOut &) |
| Save the base classes (with save_data_state) and the members in the same order that the StateIn CTOR initializes them. More... | |
| RefSCDimension | dim_natom3 () |
| Returns a smart reference to an SCDimension equal to the number of atoms in the molecule times 3. | |
| Ref<Molecule> | molecule () const |
| Returns the molecule. | |
| virtual void | print (std::ostream &=ExEnv::out()) const=0 |
| Print the coordinate. | |
| virtual void | print_simples (std::ostream &=ExEnv::out()) const=0 |
| virtual RefSCDimension | dim ()=0 |
| Returns a smart reference to an SCDimension equal to the number of coordinates (be they Cartesian, internal, or whatever) that are being optimized. | |
| int | to_cartesian (const RefSCVector &internal) |
| Given a set of displaced internal coordinates, update the cartesian coordinates of the Molecule contained herein. More... | |
| virtual int | to_cartesian (const Ref< Molecule > &mol, const RefSCVector &internal)=0 |
| virtual int | to_internal (RefSCVector &internal)=0 |
| Fill in the vector ``internal'' with the current internal coordinates. More... | |
| virtual int | to_cartesian (RefSCVector &cartesian, RefSCVector &internal)=0 |
| Convert the internal coordinate gradients in ``internal'' to Cartesian coordinates and copy these Cartesian coordinate gradients to ``cartesian''. More... | |
| virtual int | to_internal (RefSCVector &internal, RefSCVector &cartesian)=0 |
| Convert the Cartesian coordinate gradients in ``cartesian'' to internal coordinates and copy these internal coordinate gradients to ``internal''. More... | |
| virtual int | to_cartesian (RefSymmSCMatrix &cartesian, RefSymmSCMatrix &internal)=0 |
| Convert the internal coordinate Hessian ``internal'' to Cartesian coordinates and copy the result to ``cartesian''. More... | |
| virtual int | to_internal (RefSymmSCMatrix &internal, RefSymmSCMatrix &cartesian)=0 |
| Convert the Cartesian coordinate Hessian ``cartesian'' to internal coordinates and copy the result to ``internal''. More... | |
| virtual void | guess_hessian (RefSymmSCMatrix &hessian)=0 |
| Calculate an approximate hessian and place the result in ``hessian''. | |
| virtual RefSymmSCMatrix | inverse_hessian (RefSymmSCMatrix &)=0 |
| Given an Hessian, return the inverse of that hessian. More... | |
| virtual int | nconstrained () |
| Returns the number of constrained coordinates. | |
| virtual Ref<NonlinearTransform> | change_coordinates () |
| When this is called, MoleculeCoor may select a new internal coordinate system and return a transform to it. More... | |
| Ref<SCMatrixKit> | matrixkit () const |
Protected Attributes | |
| Ref<Molecule> | molecule_ |
| RefSCDimension | dnatom3_ |
| Ref<SCMatrixKit> | matrixkit_ |
| int | debug_ |
It is used to convert a molecule's cartesian coordinates to and from this coordinate system.
|
|
The KeyVal constructor.
|
|
|
When this is called, MoleculeCoor may select a new internal coordinate system and return a transform to it. The default action is to not change anything and return an IdentityTransform. Reimplemented in SymmMolecularCoor. |
|
|
Given an Hessian, return the inverse of that hessian. For singular matrices this should return the generalized inverse. Reimplemented in SymmMolecularCoor, RedundMolecularCoor, and CartMolecularCoor. |
|
|
Save the base classes (with save_data_state) and the members in the same order that the StateIn CTOR initializes them. This must be implemented by the derived class if the class has data. Reimplemented from SavableState. Reimplemented in IntMolecularCoor, SymmMolecularCoor, RedundMolecularCoor, and CartMolecularCoor. |
|
|
Convert the internal coordinate Hessian ``internal'' to Cartesian coordinates and copy the result to ``cartesian''. Only the variable internal coordinate force constants are transformed. Reimplemented in IntMolecularCoor, and CartMolecularCoor. |
|
|
Convert the internal coordinate gradients in ``internal'' to Cartesian coordinates and copy these Cartesian coordinate gradients to ``cartesian''. Only the variable internal coordinate gradients are transformed. Reimplemented in IntMolecularCoor, and CartMolecularCoor. |
|
|
Given a set of displaced internal coordinates, update the cartesian coordinates of the Molecule contained herein. This function does not change the vector ``internal''. |
|
|
Convert the Cartesian coordinate Hessian ``cartesian'' to internal coordinates and copy the result to ``internal''. Only the variable internal coordinate force constants are calculated. Reimplemented in IntMolecularCoor, and CartMolecularCoor. |
|
|
Convert the Cartesian coordinate gradients in ``cartesian'' to internal coordinates and copy these internal coordinate gradients to ``internal''. Only the variable internal coordinate gradients are calculated. Reimplemented in IntMolecularCoor, and CartMolecularCoor. |
|
|
Fill in the vector ``internal'' with the current internal coordinates. Note that this member will update the values of the variable internal coordinates. Reimplemented in IntMolecularCoor, and CartMolecularCoor. |