


Next: Translation between NAMD and
Up: Additional Simulation Parameters
Previous: Alchemical Free Energy Perturbation
Contents
Subsections
Locally Enhanced Sampling
Locally enhanced sampling (LES) [16,17,18]
increases sampling and transition rates
for a portion of a molecule by the use of multiple non-interacting copies
of the enhanced atoms. These enhanced atoms experience a nonbonded
(electrostatics and van der Waals) potential that is divided by the number
of copies present; the bonded potential is not affected. In this way the
enhanced atoms can occupy the same space, while the multiple instances and
reduces barriers imposed by the nonbonded interactions increase transition
rates.
To use LES, the structure and coordinate input files must be modified to
contain multiple copies of the enhanced atoms. psfgen provides the
multiply command for this purpose. NAMD supports a maximum of 15
copies, which should be sufficient. You may create more copies than you
intend to use in a every simulation and NAMD will scale the nonbonded
potential of these atoms to zero.
Begin by generating the complete molecular structure and guessing
coordinates as described in Sec. 4. As the last
operation in your script, prior to writing the psf and pdb files, add
the multiply command, specifying the number of copies desired and
listing segments, residues, or atoms to be multiplied. For example,
multiply 4 BPTI:56 BPTI:57 will create four copies of the last
two residues of segment BPTI. You must include all atoms to be
enhanced in a single multiply command in order for the bonded
terms in the psf file to be duplicated correctly. Calling multiply
on connected sets of atoms multiple times will produce unpredictable
results, as may running other commands after multiply.
The enhanced atoms are duplicated exactly in the structure--they have
the same segment, residue, and atom names. They are distinguished only
by the value of the B (beta) column in the pdb file, which is 0 for
normal atoms and varies from 1 to the number of copies created for
enhanced atoms. The enhanced atoms may be easily observed in VMD with
the atom selection beta != 0.
In practice, LES is a simple method used to increase sampling;
no special output is generated.
The following parameters are used to enable LES:
- les
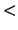 is locally enhanced sampling active?
is locally enhanced sampling active? 
Acceptable Values: on or off
Default Value: off
Description: Specifies whether or not LES is active.
- lesFactor
 number of LES images to use
number of LES images to use 
Acceptable Values: positive integer equal to or less than the number of images present
Description: This should generally be equal to the factor used in multiply
when creating the structure. The nonbonded potential of images is
divided by lesFactor. If atoms are present with an lesCol
value in lesFile that is smaller than lesFactor, then the
nonbonded potential of these atoms is set to zero.
- lesFile
 PDB file containing LES flags
PDB file containing LES flags 
Acceptable Values: UNIX filename
Default Value: coordinates
Description: PDB file to specify the LES image number of each atom.
If this parameter is not specified, then
the PDB file containing initial coordinates specified by
coordinates is used.
- lesCol
 column of PDB file containing LES flags
column of PDB file containing LES flags 
Acceptable Values: X, Y, Z, O, or B
Default Value: B
Description: Column of the PDB file to specify the LES image number of each atom.
This parameter may specify any of the floating point fields of the PDB file,
either X, Y, Z, occupancy, or beta-coupling (temperature-coupling).
A value of 0 in this column indicates that the atom is not enhanced.
Any other value should be a positive integer less than 16.
The parameter lesFactor may be varied between simulations to
interpolate between full enhancement and normal simulation (although
multiple bonded images are present at all times). If lesFactor
is decreased, the images with flags greater than lesFactor will
be decoupled from nonbonded terms, sample based only on bonded terms,
and should therefore be excluded from analysis. When increasing
lesFactor the coordinates of these abandoned images should be
reset to that of another image to avoid any bad initial contacts and
the resulting instability in the simulation.



Next: Translation between NAMD and
Up: Additional Simulation Parameters
Previous: Alchemical Free Energy Perturbation
Contents
namd@ks.uiuc.edu