




Next: Requirements
Up: Distance Restraints
Previous: Distance Restraints
- NOE { <noe-statement> } END
- is invoked from the main
level of X-PLOR.
- <noe-statement>:==
-
- ASSIgn
-
<selection> <selection> <real> <real> <real>
adds a distance restraint between the two
selected sets of atoms to the NOE database.
The interpretation of the real numbers depends on the
particular type of restraining function (see Section
18.3). For the POTEntial=BIHA, SQUA, and SOFT
options
the <real> numbers are d,
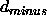 , and
, and 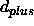 (see also Section 18.4) (default: none).
(see also Section 18.4) (default: none).
- ASYMptote
- <*class*> <real> is the slope c of the
asymptote of the soft-square function
(Eq. 18.10) (default: 0).
- AVERaging
- <*class*> R-6 | R-3 | SUM | CENTer
specifies the averaging method (Section 18.2)
(default: R-6).
- BHIG <*class*> <real>
- specifies the height of the
potential barriers in the high dimensional restraining function
(Eq. 18.15),
only for POTEntial=HIGH (default: 0.01).
- CEILing=<real>
- specifies the ceiling value for energy constants;
it applies to most restraining functions
(Section 18.3) (default: 30).
- CLASsification <class-name>
- partitions the distance restraints
database; it applies
to all subsequent ASSIgn entries until another CLASS entry is issued
(default: NIL).
- COUNtviol <class>
- counts violations in class and
stores the smallest violation in the symbol ``$RESULT";
action is taken when this statement is issued.
- DISTribute <class1> <class2> <real>
- distributes
distance restraints between two classes: the
restraint will be put in <class1> if the distance
is less than r (the specified real number);
it will be put in <class2> if the
distance is greater than or equal to r.
Action is taken only when this statement is issued.
- MONOmers
- <*class*> <integer> specifies the
number of monomers for the AVERage=SUM option
(Section 18.2) (default: 1).
- NCOUnt
- <*class*> <integer> specifies
number of assign statements for the high dimensional restraining
function (Eq. 18.15); only even values are
allowed (default: 2).
- NREStraints=<integer>
- is a required parameter that specifies
the maximum expected number of distance restraints. It
has to be greater than
or equal to the actual number of restraints. The parameter is used to
allocate space dynamically for the restraint list (default: 200).
- POTEntial
- <*class*>
BIHArmonic | SQUAre | SOFTsquare | SYMMetry | HIGH | 3DPO
specifies the restraining function (Section 18.3)
(default: BIHArmonic).
- PREDict
- { <predict-statement> } END can be used to
predict interproton distances based on the Cartesian coordinates in the
MAIN coordinate set. It
lists all distances between the FROM and the TO selections of
atoms that are within the range CUTOn ... CUTOff. Distances
are
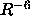 averaged over the GROUps that are
defined in the topology file for each residue
(see Section 3.1.1).
Current distance restraint information is overwritten by this statement.
averaged over the GROUps that are
defined in the topology file for each residue
(see Section 3.1.1).
Current distance restraint information is overwritten by this statement.
- PRINt THREshold=<real>
- prints distance violations
greater than the specified real number. This statement also
declares two symbols, $RESULT and $VIOLATIONS, which correspond to the
rms deviation of the distances and the number of violations greater than
the THREshold value, respectively.
The definition of the deviation depends on which restraining function
is specified: in the case of the SQUAre-well and SOFTsquare
functions, the deviation refers to the difference
between the actual distance and the upper or lower
bound if the distance is outside the error range; in
the case of the BIHArmonic function,
the deviation refers to the difference between the actual
distance and the target distance.
(A more precise definition is
given by
 in Section 18.3.)
in Section 18.3.)
- RAVErage
- <*class*>
{ <raverage-statement> } END
This statement is used to compute a running-average
of each interproton distance. When the running average
is turned on, an average distance is accumulated for each
interatomic distance defined within the class of noe restraints.
This running-average is updated at each time-step in molecular
dynamics, and at each step in energy minimization. This facility
is described in more detail in Section 18.11.
- RESEt
- erases the current NOE database.
- RSWItch <*class*> <real>
- is the switch parameter
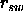 between
the square-well and the asymptote (Eq. 18.10)
(default: 10).
between
the square-well and the asymptote (Eq. 18.10)
(default: 10).
- SCALe <*class*> <real>
- scales the restraint
class (Section 18.3) (default: 1).
- SOEXponent <*class*> <real>
- is an exponent;
it is for the soft-square
function (Eq. 18.10) (default: 2).
- SQCOnstant <*class*> <real>
- is an additional scale factor;
it is for the square-well or soft-square function
(Eqs. 18.8 and 18.10) (default: 20).
- SQEXponent <*class*> <real>
- is an exponent;
it is for the square-well
or soft-square function (Eqs. 18.8
and 18.10) (default: 2).
- SQOFfset <*class*> <real>
- applies
a negative offset value to all upper bounds
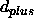 of the
specified class(es). This offset is
applied to the square-well or soft-square functions
(Eqs. 18.8 and 18.10) and to the
bounds used by distance geometry (see Chapter 19) (default: 0).
of the
specified class(es). This offset is
applied to the square-well or soft-square functions
(Eqs. 18.8 and 18.10) and to the
bounds used by distance geometry (see Chapter 19) (default: 0).
- TAVErage
- <*class*> { <taverage-statement> } END
This statement is used to turn on or off time-averaged noe restraints.
This facility
is described in more detail in Section 18.11.
- TEMPerature=<real>
- is the temperature in K; it is
only for the biharmonic function (Eq. 18.6)
(default: 300).
- <predict-statement>:==
-
- CUTOff=<real>
- specifies an upper limit for acceptable interproton
distances (default: 10.0 Å).
- CUTOn=<real>
- specifies a lower limit for acceptable interproton
distances (default: 0.0 Å).
- FROM=<selection>
- specifies an atom selection (default: none).
- TO=<selection>
- specifies an atom selection (default: none).
- <raverage-statement>:==
-
- ON
- Turns on the running-average.
- OFF
- Turns off the running-average.
- EXPOnent=<integer>
- Specifies the exponent used in
distance--averaging (default: 3).
- RESEt=<CURRent,CONStraint>
- Resets the
distance running-averages setting the new initial values
to either the current distances (CURRent) or the
target distances in the NOE database (CONStraint).
- <taverage-statement>:==
-
- ON
- Turns on time-averaged restraints.
- OFF
- Turns off time-averaged restraints.
- TAU=<integer>
- Specifies the exponential decay
constant (in number of time steps) used in the averaging
(default: 1000 time steps).
- EXPOnent=<integer>
- Specifies the exponent used in
distance--averaging (default: 3).
- FORCe=<CONServative,NONConservative>
- Specifies
whether a conservative
or non-conservative force is used in time-averaging
(default: CONServative).
- RESEt=<CURRent,CONStraint>
- Resets the
distance time-averages setting the new initial values
to either the current distances (CURRent) or the
target distances in the NOE database (CONStraint).





Next: Requirements
Up: Distance Restraints
Previous: Distance Restraints
Web Manager
Sat Mar 11 09:37:37 PST 1995
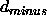 , and
, and 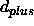 (see also Section 18.4) (default: none).
(see also Section 18.4) (default: none).
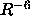 averaged over the GROUps that are
defined in the topology file for each residue
(see Section 3.1.1).
Current distance restraint information is overwritten by this statement.
averaged over the GROUps that are
defined in the topology file for each residue
(see Section 3.1.1).
Current distance restraint information is overwritten by this statement.
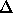 in Section 18.3.)
in Section 18.3.)
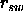 between
the square-well and the asymptote (Eq. 18.10)
(default: 10).
between
the square-well and the asymptote (Eq. 18.10)
(default: 10).
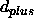 of the
specified class(es). This offset is
applied to the square-well or soft-square functions
(Eqs. 18.8 and 18.10) and to the
bounds used by distance geometry (see Chapter 19) (default: 0).
of the
specified class(es). This offset is
applied to the square-well or soft-square functions
(Eqs. 18.8 and 18.10) and to the
bounds used by distance geometry (see Chapter 19) (default: 0).