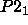 (Strong 1990), with the
b-axis unique, and a non-crystallographic twofold symmetry is
present. The following strategy was employed to
solve the structure:
(Strong 1990), with the
b-axis unique, and a non-crystallographic twofold symmetry is
present. The following strategy was employed to
solve the structure:
The following sections show how the 26-10 Fab fragment complexed
with digoxin was solved by generalized molecular
replacement (Brünger 1991c).
The space group of the 26-10 Fab/digoxin crystals
is 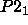 (Strong 1990), with the
b-axis unique, and a non-crystallographic twofold symmetry is
present. The following strategy was employed to
solve the structure:
(Strong 1990), with the
b-axis unique, and a non-crystallographic twofold symmetry is
present. The following strategy was employed to
solve the structure:
 apart) and repeat steps 3--6 until a good solution is found.
This strategy is also applicable to other multidomain proteins
or PC-refinements of other degrees of freedom. The
modification of the Fab example input files should be straightforward.
An overview of the strategy is shown in Fig. 17.1.
apart) and repeat steps 3--6 until a good solution is found.
This strategy is also applicable to other multidomain proteins
or PC-refinements of other degrees of freedom. The
modification of the Fab example input files should be straightforward.
An overview of the strategy is shown in Fig. 17.1.
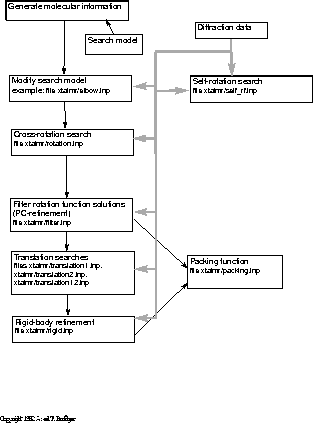
Figure 17.1: Overview of molecular replacement.