This facility can be used to obtain bond, bond angle, dihedral angle, or improper angle equilibrium parameters and energy constants from selected atoms of Cartesian coordinate sets. The coordinates are specified in the main coordinate set. The statement learns parameters only for those interaction terms that are turned on by the flags statement (Section 4.5 ). The learned parameters will take precedence over the type-based parameters.
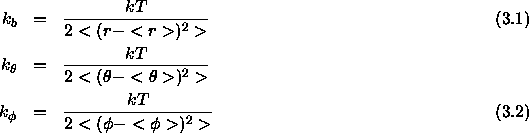
The equilibrium geometry parameters can be directly obtained from a single coordinate set or averaged over successive coordinate sets. If just a single coordinate set is available, one can learn only the equilibrium geometry, not the energy constants. If an ensemble of coordinates is available, energy constants can be derived assuming equipartition of energy among the different internal coordinates. This is only approximately verified in a real system, since there is actually coupling among the internal coordinates.
The brackets represent an average over the ensemble of coordinate sets.
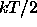 is the mean thermal energy per harmonic degree of freedom at
T=298K;
Eqs. 4.4, 4.5, and 4.6
define the other constants.
The last expression assumes that all dihedral angles and torsion
angles are represented by a harmonic functional form with
periodicity n set to zero (Eq. 4.6). In fact,
the learn facility will set the periodicity of all ``learned"
dihedral and improper angles to zero. In the case that one of the
variances in the denominators of Eq. 3.1 becomes zero, the
corresponding energy constant is set to 999999.
is the mean thermal energy per harmonic degree of freedom at
T=298K;
Eqs. 4.4, 4.5, and 4.6
define the other constants.
The last expression assumes that all dihedral angles and torsion
angles are represented by a harmonic functional form with
periodicity n set to zero (Eq. 4.6). In fact,
the learn facility will set the periodicity of all ``learned"
dihedral and improper angles to zero. In the case that one of the
variances in the denominators of Eq. 3.1 becomes zero, the
corresponding energy constant is set to 999999.
Parameter learning is not possible for nonbonded and hydrogen-bonded parameters.
learn initiate selection=( name c* ) MODE=STATistics end learn accumulate end learn terminate end